SPSS is a data mining and statistical analysis software. Resent versions (v14 - v17) allow to use Python to control SPSS. This can be quite handy, especially if there are lots of variables and lots of data files to analyze and such analysis is often repeated. In such a case, writing some script in Python that can execute SPSS analysis automatically can save you lots of time. This is the first post out of three, which will present some example of using Python and SPSS together. First, let me present what soft and hardware I have:
1. Start SPSS.
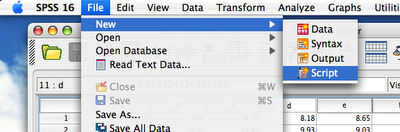
2. Create new script (File->New->Script):
- Intel Mac X 10.4.11
- SPSS 16.0.1 with SPSS-Python Integration Plug-In
- Python 2.5
Check if SPSS-Python Integration Plug-In works
Before we can proceed any further we have to check if SPSS-Python Integration Plug-In is working, and we can control SPSS using Python. To do this I usually use the following procedure:1. Start SPSS.
2. Create new script (File->New->Script):
 This results in a python IDLE console:
This results in a python IDLE console: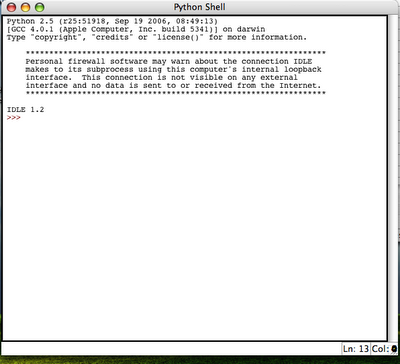 3. Try to import spss library and execute some simple spss function (e.g. spss.GetVariableCount()):
3. Try to import spss library and execute some simple spss function (e.g. spss.GetVariableCount()):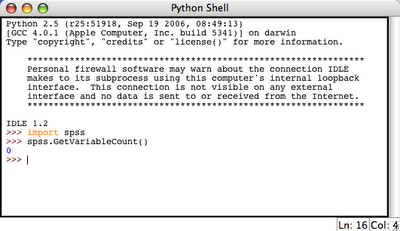 This resulted in 0 which is OK, because we do not have any active dataset.
This resulted in 0 which is OK, because we do not have any active dataset.So it appears that SPSS-Python union is working. Therefore, we can do something useful now.
Note
More information on SPSS-Python programming along with examples can be found in SPSS Programming and Data Management.






