| function plotDiff2D() %p. 351, eg. 1 t=[-2:0.1:2]; cT=[-2:.5:2]; for c1=cT for c2=cT x1=c1*exp(3*t)+c2*exp(-t); x2=c1*2*exp(3*t)-2*c2*exp(-t); lineS='b-'; if c1==0 || c2==0, lineS='r-'; end hold on; plot3(x1,x2,t,lineS); end end limm=6 xlim([-limm limm]);ylim([-limm limm]); %axis square xlabel('x1'); ylabel('x2'); zlabel('t'); grid on; |
i.e. some stuff and junk about Python, Perl, Matlab, Ruby, Mac X, Linux, Solaris, ...
Monday, January 14, 2008
Thursday, January 10, 2008
Octave 3: first impression
 Whats more, the problems with eigenvalues that I had with Python (see this post) does not exist in Octave 3. I have to say, that for now I have been working only with eigenvalues, inverse matrices, and multiplication of matrices. But I am encourge to use Octave 3 for longer, to other tasks, and we will see what will happen.
Whats more, the problems with eigenvalues that I had with Python (see this post) does not exist in Octave 3. I have to say, that for now I have been working only with eigenvalues, inverse matrices, and multiplication of matrices. But I am encourge to use Octave 3 for longer, to other tasks, and we will see what will happen.Update:
I found problem with 3D plots. When you draw such plot, often the plot window freezes. As a result is not possible to rotate, zoom or do any operations on the figure. Its very annoying. I use gnuplot as plot engine. I think I will try to use the other one. During installation of the Octave 3 on Windows in one point the user is asked to choose an engine plot. I should try the second one, and we will see what will happen.
Wednesday, January 09, 2008
Python: eigenvalues with Scipy and Sympy
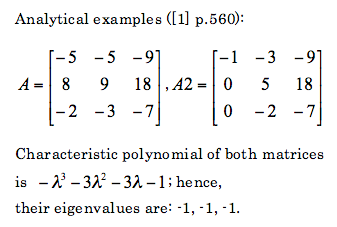
Lets see what does Python calculate:
Scipy
from scipy import *
from scipy.linalg import *
A=matrix([ [-5,-5,-9],[8,9,18],[-2,-3,-7] ])
print eigvals(A)
# [-0.999989 +1.90461984e-05j -0.999989 -1.90461984e-05j
# -1.00002199 +0.00000000e+00j]
A2=matrix([ [-5,-5,-9],[8,9,18],[-2,-3,-7] ])
print eigvals(A2)
# [-1.+0.j -1.00000005+0.j -0.99999995+0.j]Sympy
from sympy import *
from sympy.matrices import Matrix
x=Symbol('x')
A=Matrix(( [-5,-5,-9],[8,9,18],[-2,-3,-7] ))
print roots(A.charpoly(x),x)
# Returns error!!!
A2=Matrix(( [-1,-3,-9],[0,5,18],[0,-2,-7] ))
print roots(A2.charpoly(x),x)
# Returns error!!!
Based on these to examples it is clearly seen that these two packaged are unreliable, at least as far as calculation of eigenvalues is considered.
For matrices 2x2 it was noticed that the eigenvalues are correctly calculated both for real and complex values.
For comparison lets check what does Matlab returns:
A=[-5,-5,-9;8,9,18;-2,-3,-7]
eig(A)
ans =
-1.0000 + 0.0000i
-1.0000 - 0.0000i
-1.0000
A2=[-5,-5,-9;8,9,18;-2,-3,-7]
eig(A2)
ans =
-1.0000 + 0.0000i
-1.0000 - 0.0000i
-1.0000
So it is seen that Matlab does correctly calculate these eigenvalues.
Even if one manually calculates characteristic polynomial and would like to gets roots of it, I would recommend neither Scipy or Sympy. As an example I calculate roots of the previously obtained characteristic polynomial for matrices A and A2:
import scipy as sc
p=sc.poly1d([-1,-3,-3,-1])
print p= sc.roots(p)
# [-1.0000086 +0.00000000e+00j -0.9999957 +7.44736442e-06j
# -0.9999957 -7.44736442e-06j]
import sympy as sy
x=sy.Symbol('x')
print sy.solve(-x**3-3*x**2-3*x-1==0,x)
#[-1]
As can be seen calculation of roots of polynomials is also not good. In the second case there is one root (-1) instead of three (-1,-1,-1).
For comparison lets check Matlab (or Octave 3):
p=[-1,-3,-3,-1]
roots(p)
ans =
-1.0000
-1.0000 + 0.0000i
-1.0000 - 0.0000i
Based on these results it is seen that its better to use Matlab for calculation of eignevalues.
References:
[1] S.I. Grossman, Multivariable calculus, linear algebra and differential equation, Second edition, 1986
Sunday, December 02, 2007
Python Imaging Library - not working with 16 bit images


#!/usr/bin/env python
'''
Convert tif images from Ludvig 2008.
This scripts take all tifs in input dir, and changes
tif files into tiff. Additionali it takes
right knee x-rays and flips them horizontally,
to have all x-ray in the same format.
'''
#from myUtil import *
import re
import os
import Image
import Tkinter, tkFileDialog, os.path
from scipy import *
def flip_horizontally(inDir,inFile,outFile=None):
imgpath=inDir+inFile
im = Image.open(imgpath)
out = im.transpose(Image.FLIP_LEFT_RIGHT)
if outFile is None: outFile=inFile
base=os.path.splitext(outFile)[0]
out.save(inDir+base+'_flopped.tiff')
def main():
inDir=myGetDir('./')
print inDir
files=os.listdir(inDir)
#change_file_ext(inDir,r'\.tif+$','.tiff')
for f in files:
if f.find('tiff')==-1: continue
if f.find('test')==-1: continue
print f
flip_horizontally(inDir,f,f)
def myGetDir(indir='./',putTitle='Select dir'):
"""Get one dir name"""
root = Tkinter.Tk()
root.withdraw()
dirr=tkFileDialog.askdirectory(initialdir=indir,
title=putTitle)
if len(dirr)==0: exit(1)
return dirr+'/'
if __name__ == '__main__':
main()
The result of the above code is:


It is clearly seen that flopping 16-bit image gives wrong results.
Thursday, November 29, 2007
Friday, November 16, 2007
Matlab: Easter egg - spy
 Interesting, isn't it:-)
Interesting, isn't it:-)
Monday, August 27, 2007
Monday, July 23, 2007
PSNR (Peak Signal-to-Noise Ratio)
Compressing an image is significantly different than compressing raw binary data. Of course, general purpose compression programs can be used to compress images, but the result is less than optimal. This is because images have certain statistical properties which can be exploited by encoders specifically designed for them. Also, some of the finer details in the image can be sacrificed for the sake of saving a little more bandwidth or storage space. This also means that lossy compression techniques can be used in this area.
Lossless compression involves with compressing data which, when decompressed, will be an exact replica of the original data. This is the case when binary data such as executables, documents etc. are compressed. They need to be exactly reproduced when decompressed. On the other hand, images (and music too) need not be reproduced 'exactly'. An approximation of the original image is enough for most purposes, as long as the error between the original and the compressed image is tolerable.
Error Metrics
Two of the error metrics used to compare the various image compression techniques are the Mean Square Error (MSE) and the Peak Signal to Noise Ratio (PSNR). The MSE is the cumulative squared error between the compressed and the original image, whereas PSNR is a measure of the peak error. The mathematical formulas for the two are
MSE =
PSNR = 20 * log10 (255 / sqrt(MSE))
where I(x,y) is the original image, I'(x,y) is the approximated version (which is actually the decompressed image) and M,N are the dimensions of the images. A lower value for MSE means lesser error, and as seen from the inverse relation between the MSE and PSNR, this translates to a high value of PSNR. Logically, a higher value of PSNR is good because it means that the ratio of Signal to Noise is higher. Here, the 'signal' is the original image, and the 'noise' is the error in reconstruction. So, if you find a compression scheme having a lower MSE (and a high PSNR), you can recognize that it is a better one.
Monday, June 25, 2007
Matlab: Mandelbrot set
function mandelFrac
% MATLAB and Octave code to generate
%a Mandelbrot fractal
% Number of points in side of image and
% number of iterations in the Mandelbrot
% fractal calculation
npts=1000;
niter=51;
% Generating z = 0 (real and
% imaginary part)
zRe=zeros(npts,npts);
zIm=zeros(npts,npts);
% Generating the constant k (real and
% imaginary part)
kRe=repmat(linspace(-1.5,0.5,npts),npts,1);
kIm=repmat(linspace(-1,1,npts)',1,npts);
% Iterating
for j=1:niter
% Calculating q = z*z + k in complex space
% q is a temporary variable to store the result
qRe=zRe.*zRe-zIm.*zIm+kRe;
qIm=2.*zRe.*zIm+kIm;
% Assigning the q values to z constraining between
% -5 and 5 to avoid numerical divergences
zRe=qRe;
qgtfive= find(qRe > 5.);
zRe(qgtfive)=5.;
qltmfive=find(qRe<-5.);
zRe(qltmfive)=-5.;
zIm=qIm;
hgtfive=find(qIm>5.);
zIm(hgtfive)=5.;
hltmfive=find(qIm<-5.);
zIm(hltmfive)=-5.;
end
% Lines below this one are commented out when making
% the benchmark.
% Generating plot
% Generating the image to plot
ima=log( sqrt(zRe.*zRe+zIm.*zIm) + 1);
% Plotting the image
imagesc(ima);



