CPPFLAGS="-I/usr/include/tirpc" ./configure --prefix=/opt/python362 --enable-shared -I/usr/include/tirpc
i.e. some stuff and junk about Python, Perl, Matlab, Ruby, Mac X, Linux, Solaris, ...
Showing posts with label Python. Show all posts
Showing posts with label Python. Show all posts
Tuesday, October 02, 2018
Arch: Python 3.6 rpc/rpc.h missing
The solution was to manually specify where `rpc/rpc.h` was for configuration:
Friday, October 23, 2015
Make python 3 virtual envirnoment on Ubuntu 15.10
sudo apt-get install python3-venv
python3 -m venv ~/mypy34env/
source ~/mypy34env/bin/activate
And then install python packages using pip, for example, pip install ipython
Thursday, July 03, 2014
How to install virtual environment of Python 3.4.x from source in Ubuntu 14.04
Below is a recipe that I use to used to install and set-up virtual environment of Python 3.4.x in Ubuntu 14.04 x64.
Thursday, June 26, 2014
Reading DICOM files in Python 3
In Python 3, dicom files can be read, analysied and processed using Wand which is Python binding for ImageMagic. The example below shows how to get medical and image date embedded in DICOM and also how to disply the image itself.
Labels:
imagemagick,
Python,
Wand
Python 3.4 and Pillow 2.4 with JPEG2000 (openjpeg 2) support in Ubuntu 14.04
Pillow support for JPEG2000 comes from openjpeg 2 library. Unfortunatly, Ubuntu's libopenjpeg2 package is not version 2 of the openjpeg library, but actually version 1.3 which wont work with Pillow.
Installing Pillow 2.4 (pip install -I pillow) simply results in "OPENJPEG (JPEG2000) support not available".
Thus, it is needed to compile the openjpeg library 2.0 from source. For this, first lets download the openjpeg 2.0.1:
Please note that we install version of openjpeg 2.0, rather than newer 2.1. Pillow wont recognize openjpeg 2.1 either.
Assuming everything went fine, we can reinstall Pillow:
If openjpeg 2.0.1 was detected successfully, we should get the following info "OPENJPEG (JPEG2000) support available" among others.
Installing Pillow 2.4 (pip install -I pillow) simply results in "OPENJPEG (JPEG2000) support not available".
Thus, it is needed to compile the openjpeg library 2.0 from source. For this, first lets download the openjpeg 2.0.1:
wget http://downloads.sourceforge.net/project/openjpeg.mirror/2.0.1/openjpeg-2.0.1.tar.gz
tar xzvf openjpeg-2.0.1.tar.gz
cd openjpeg-2.0.1/
cmake .
make
sudo make install
Please note that we install version of openjpeg 2.0, rather than newer 2.1. Pillow wont recognize openjpeg 2.1 either.
Assuming everything went fine, we can reinstall Pillow:
pip install -I pillowIf openjpeg 2.0.1 was detected successfully, we should get the following info "OPENJPEG (JPEG2000) support available" among others.
Friday, May 23, 2014
Python 3 and tkinter: scroll canvas with mouse wheel and drag it around with middle mouse button
This is an example in Python 3.4 running in 64-bit Ubuntu 14.04 of a canvas that can be scrolled with mouse wheel and dragged around with middle mouse button. Hope this example will be useful to you.
Thursday, June 18, 2009
python: numbers starting with zero are octagonal
I was debugging my script and I found that a number that starts from zero is different that the number without zero. This is because these numbers are octal literals. For example
In [1]: a=23
In [2]: b=023
In [3]: a==b
Out[3]: False
In [4]: print a,b
23 19It appears that when a number starts from zero it is treated as an octagonal number rather than decimal number. Good to know, as it is easy to make mistake.
Labels:
Python
Friday, May 22, 2009
python: simple start with simplejson
JSON (i.e. JavaScript Object Notation) is a popular way of exchanging data in a string format. It can also be used to provide setup parameters for a program. The second option I used to show very basic JSON usage in python with simplejson package. Lets assume that we have a txt file called data.json with the following JSON code:
{
"Name": "W",
"Job": "Programmer",
"Movies": ["Star Wars", "Star Trek", "Terminator"],
"Address" : {
"Street": "Some Streat",
"House": 3,
"City": "Some City"
}
}
The above JSON code can be read into python object using load function as followsIn [7]: import simplejson as json
In [8]: f=open("data.json")
In [9]: data=json.load(f)
In [10]: f.close()
In [11]: data
Out[11]:
{'Address': {'House': 3, 'Street': 'Some Streat', 'City': 'Some City'},
'Job': 'Programmer',
'Movies': ['Star Wars', 'Star Trek', 'Terminator'],
'Name': 'W'}
In [12]: print data['Address']['House']
3
In [13]:
That's it:-)
Labels:
Python
Monday, May 04, 2009
Matlab 2008b: Execute matlab from python in CentOS 5.2
I have Matlab 2008b running on CentOS 5.2 server. I wanted to be able to execute Matlab commands from python. After a search I found mlabwrap 1.0.1. I decided to install it. On CentOS 5.2 I have python 2.4.3. I also installed numpy 1.0.4 from epel repository. numpy is required to install mlabwrap. To install mlabwrap I fallowed the instructions. Fortunately, I had no problems with the installation.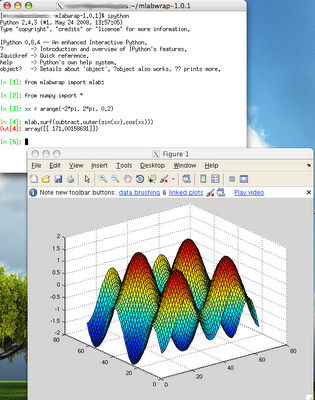 Note: the appearance of the windows in the image is from Mac X because I X forward them from CentOS server.
Note: the appearance of the windows in the image is from Mac X because I X forward them from CentOS server.
 Note: the appearance of the windows in the image is from Mac X because I X forward them from CentOS server.
Note: the appearance of the windows in the image is from Mac X because I X forward them from CentOS server.
Thursday, March 26, 2009
Arch linux: very first impression
Usually I don't write any reviews or any opinions on linux distros. However, I have to make an exception. The reason is that I have just install Arch linux! Arch linux is 
a lightweight and flexible Linux® distribution that tries to Keep It Simple.First on virtual box though, just to see what all the fuss is about. What I see is that this distro is quite interesting, it was easy to install. It was easy to set up fluxbox, and some other programs that I need (e.g. python-numpy, python-scipy, matplotlib). Currently I’m using CentOS on my HPC (high performance computing) server, and ubuntu 8.10 on desktop. In few weeks Ubuntu 9.04 released and I strongly think that I may move to Arch on desktop! Not 100% sure now, but I keep testing arch on virtualbox. Definitely, the first impression is positive! If I'll have some further info about this, I will update this post.
Monday, March 16, 2009
LS-DYNA parallel: SMP vs MPP
LS-DYNA, is an part of ANSYS software and it is a general purpose transient dynamic finite element program capable of simulating complex real world problems.
For large models, you can use the shared memory parallel processing (SMP) or the massively parallel processing (MPP) capabilities of ANSYS LS-DYNA to shorten the elapsed time necessary to run an analysis.
The shared memory parallel processing capabilities allow you to distribute model-solving power over multiple processors on the same machine.
The massively parallel processing (MPP) capabilities of ANSYS LS-DYNA allow you to run the LS-DYNA solver over a cluster of machines or use multiple processors on a single machine.
My question is which one is faster and how faster?
To answer to this question, I performed LS-DYNA simulation of an official LS-DYNA example airbag.deploy.k, once with SMP and once with MPP for different number of CPUs. To use LS-DYNA SMP I used the following example command:ls971.e110 pr=aa_r_dy ncpu=%d i=airbag.deploy.k where pr=aa_r_dy is my license, and %d is the number of CPUs to be used for solving the airbag problem. For LS-DYNA MPP I used:lsdyna110 -dis -np %d pr=aa_r_dy i=.airbag.deploy.k where %d is the number of CPUs to be used.The above commands were used in a short Python script (it is here) that executes them with different values of %d, calculates execution times, and generates file with the time results. The times obtained were plotted against number of CPUs used:
 The tests were executed on a single server with 4 Intel Xeon processors and 24GB RAM. Each processor had 6 cores. The operating system (CentOS 5.2) sees this as 24 CPUs.
The tests were executed on a single server with 4 Intel Xeon processors and 24GB RAM. Each processor had 6 cores. The operating system (CentOS 5.2) sees this as 24 CPUs.Conclusions
It can be seen for the graph, that solving LS-DYNA problems using MPP is faster than using SMP. However, the execution time does not reduce linearly with the number of CPUs. One reason may be that airbag.deploy.k is not well suited for solving using large number of CPUs.Sunday, January 25, 2009
Python: Example of Threads and Queue
Just an example of making thread workers, that executes tasks from the task queue. I used it to check if a PC box with multicore processor is really 100% busy with threads. The code multiplies some large, random matrices, so it does not do anything useful. However, it can serve as an example or a template of a simple multithread application.
import threading, Queue, time
from scipy import *
class MultiplyMatrixes(threading.Thread):
def __init__(self,queue,name):
self.queue=queue
threading.Thread.__init__(self,name=name)
def run(self):
while True:
#wait until there is a task
# in queue. Get task from gueue.
n=self.queue.get()
print "Spawning: ",self.getName(),n
if n !=None:
# do the task
p=self.doMultiplication(n)
print "Finished:", self.getName(),n,p
#indicate that the task has been
#complited
self.queue.task_done()
def doMultiplication(self,n):
m1=matrix(rand(n,n)) #some random matrix
m2=matrix(rand(n,n)) #some random matrix
m3=m1*m2
return m3.mean() # return mean
class MainThread():
def __init__(self,noOfThreads):
self.no=noOfThreads
def doTest(self):
t1=time.time()
self.test()
print "Elapsed time",time.time()-t1
def test(self):
#change maxMtxSize for longer or shorter
# execution
minMtxSize,maxMtxSize=100,1200
nT=range(minMtxSize,maxMtxSize,100)
#make task queue
queue=Queue.Queue()
#create number of workers (threads)
for i in range(self.no):
p=MultiplyMatrixes(queue,name=str(i+1))
p.setDaemon(True)
p.start()
#put some tasks into the queue
for n in nT:
queue.put(n)
#wait until the queue is finshed (no more tasks)
queue.join()
#give some time for threads
#to finished before exiting.
time.sleep(1)
#Otherwise the follwing error is more likely to occur:
# Exception in thread 1 (most likely raised during interpreter shutdown):
# Traceback (most recent call last):
# File "/usr/lib/python2.5/threading.py", line 486, in __bootstrap_inner
# File "multithreads.py", line 19, in run
# File "/usr/lib/python2.5/Queue.py", line 165, in get
# File "/usr/lib/python2.5/threading.py", line 209, in wait
# : 'NoneType' object is not callable
print "Finished!!!"
def test():
MP=MainThread(noOfThreads=4)
MP.doTest()
if __name__ == '__main__':
test() Spawning: 1 100
Finished: 1 100 25.3551569392
Spawning: 2 200
Spawning: 3 300
Spawning: 1 400
Spawning: 4 500
Finished: 2 200 49.9585540799
Spawning: 2 600
Finished: 3 300 75.163025977
Spawning: 3 700
Finished: 1 400 99.7075107712
Spawning: 1 800
Finished: 4 500 124.849660963
Spawning: 4 900
Finished: 2 600 149.99426813
Spawning: 2 1000
Finished: 3 700 175.035573614
Spawning: 3 1100
Finished: 1 800 199.893719979
Finished: 4 900 225.168134578
Finished: 2 1000 250.157828058
Finished: 3 1100 275.025345136
Finished!!!
Elapsed time 73.96342206
Labels:
Python
Python: Python Imaging Library - 16 bit images
The problem described in the previews post about 16-bit images in python can be overcome by converting image into numpy array, operating in numpy array, and finally converting array into image and saving it:
Using this function instead of the one used before produces correct results.
def flip_horizontally2(inDir,inFile,outFile=None):
''' Open 16-bit or 8-bit image, flop it horizontally,
and save it
.'''
imgpath=inDir+inFile
im = Image.open(imgpath)
print im.mode,im.size[1], im.size[0]
#convert image into array according to im.mode
if im.mode in ("L",):
a = fromstring(im.tostring(), uint8)
elif im.mode in ("I;16",):
a = fromstring(im.tostring(), uint16)
a.shape = im.size[1], im.size[0]
a=fliplr(a) #flip horizontally
#convert array into image according to a.dtype
if a.dtype == uint8: mode = "L"
elif a.dtype == uint16: mode = "I;16"
else:
raise ValueError, "unsupported image mode"
out=Image.fromstring(mode, (a.shape[1],
a.shape[0]), a.tostring())
#save image
if outFile is None: outFile=inFile
base=os.path.splitext(outFile)[0]
out.save(inDir+base+'_flopped2.tiff')
Using this function instead of the one used before produces correct results.
Labels:
Python
Python: an image to an array and an array to an image
Converting 8-bit gray-scale level images to numpy array, and generating a 8-bit gray-scale level image from array.
Labels:
Python
Python: check if a string is a number
To check if a string represents a number, this may be used:
Examples:
def isnum(s):
try:
float(s)
except ValueError:
return False
return True
Examples:
In [57]: isnum('abs')
Out[57]: False
In [58]: isnum('2.5')
Out[58]: True
In [59]: isnum('-2.5')
Out[59]: True
In [60]: isnum("2e-3")
Out[60]: True
In [61]: isnum("2e-3 a")
Out[61]: False
In [62]: isnum("2e-3 ")
Out[62]: True
In [63]: isnum(" 1.1e+3 ")
Out[63]: True
In [63]: isnum(" 52 ")
Out[63]: True
Labels:
Python
Sunday, December 21, 2008
SPSS: programing SPSS with Python (part 1)
SPSS is a data mining and statistical analysis software. Resent versions (v14 - v17) allow to use Python to control SPSS. This can be quite handy, especially if there are lots of variables and lots of data files to analyze and such analysis is often repeated. In such a case, writing some script in Python that can execute SPSS analysis automatically can save you lots of time. This is the first post out of three, which will present some example of using Python and SPSS together. First, let me present what soft and hardware I have:
1. Start SPSS.
2. Create new script (File->New->Script):
- Intel Mac X 10.4.11
- SPSS 16.0.1 with SPSS-Python Integration Plug-In
- Python 2.5
Check if SPSS-Python Integration Plug-In works
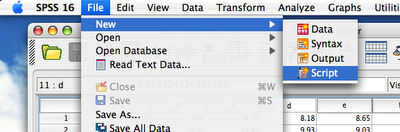
Before we can proceed any further we have to check if SPSS-Python Integration Plug-In is working, and we can control SPSS using Python. To do this I usually use the following procedure:1. Start SPSS.
2. Create new script (File->New->Script):
 This results in a python IDLE console:
This results in a python IDLE console: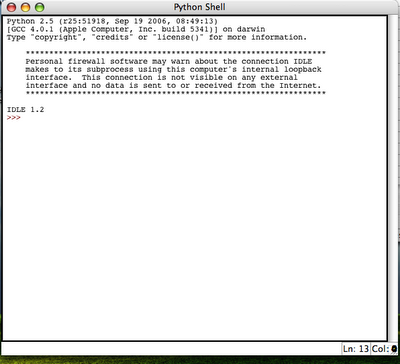 3. Try to import spss library and execute some simple spss function (e.g. spss.GetVariableCount()):
3. Try to import spss library and execute some simple spss function (e.g. spss.GetVariableCount()):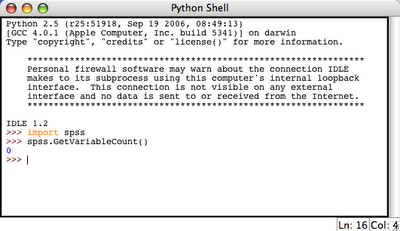 This resulted in 0 which is OK, because we do not have any active dataset.
This resulted in 0 which is OK, because we do not have any active dataset.So it appears that SPSS-Python union is working. Therefore, we can do something useful now.
Note
More information on SPSS-Python programming along with examples can be found in SPSS Programming and Data Management.
Wednesday, December 17, 2008
Python: Ellipse fitting
The algorithm below fits an ellipse into the polygon (or set of points) in such a way that the area of the ellipse fitted is equal to the area of the polygon.




from __future__ import division
from scipy import *
import pylab as p
def ellipse(ra,rb,ang,x0,y0,Nb=50):
'''ra - major axis length
rb - minor axis length
ang - angle
x0,y0 - position of centre of ellipse
Nb - No. of points that make an ellipse
based on matlab code ellipse.m written by D.G. Long,
Brigham Young University, based on the
CIRCLES.m original
written by Peter Blattner, Institute of Microtechnology,
University of
Neuchatel, Switzerland, blattner@imt.unine.ch
'''
xpos,ypos=x0,y0
radm,radn=ra,rb
an=ang
co,si=cos(an),sin(an)
the=linspace(0,2*pi,Nb)
X=radm*cos(the)*co-si*radn*sin(the)+xpos
Y=radm*cos(the)*si+co*radn*sin(the)+ypos
return X,Y
def _makexy(scale):
x=reshape(array(range(scale*2)*(scale*2)),(scale*2,scale*2))
return x,x.T
def _inside2(x,y,px,py):
'''Check if point (x,y) inside polygon deifined by (px,py).
From: http://www.ariel.com.au/a/python-point-int-poly.html
'''
poly=zip(px,py)
n = len(poly)
inside =False
p1x,p1y = poly[0]
for i in range(n+1):
p2x,p2y = poly[i % n]
if y > min(p1y,p2y):
if y <= max(p1y,p2y):
if x <= max(p1x,p2x):
if p1y != p2y:
xinters = (y-p1y)*(p2x-p1x)/(p2y-p1y)+p1x
if p1x == p2x or x <= xinters:
inside = not inside
p1x,p1y = p2x,p2y
return inside
def _polyMask(X,Y,rs):
'''Make binary mask of polygon. Values of one
in 2D array pd represents points enclosed by the polygon.'''
pd=zeros((rs*2,rs*2))
for i in range(pd.shape[0]):
for j in range(pd.shape[1]):
if _inside2(i,j,X,Y)==True:
pd[i,j]=1.0
return pd
def ellfit(X,Y,rs=50,showFig=True):
'''Fit and ellipse to a polygon defined by X,Y in such a way
that area of the ellispe fitted is the same as area of the polygon.
Based on ellfit.pro by D.G. Long, Brigham Young University.
Conditions:
1. The polygon must not be self intersecting.
2. Polygon must be closed (last point must match the first one).
3. Polygon must be centered in (0,0)
4. Polygon must be within rectangle <-1,1>x<-1,1>
'''
#Xmax,Ymax=abs(X).max(),abs(Y).max()
#X=X/Xmax
#Y=Y/Ymax
pi2=2*pi
rs2=2*rs
if showFig:
fig = p.figure(figsize=(5,5))
ax = fig.add_subplot(111)
X=X*rs+rs
Y=Y*rs+rs
xa,ya=_makexy(rs)
pd=_polyMask(X,Y,rs)
if showFig:
p.plot(X,Y,"rs-",ms=15)
for i in range(pd.shape[0]):
for j in range(pd.shape[1]):
if pd[i,j]==1.0:
pass
p.plot([i],[j],'g.',ms=10)
m00=pd.sum()
m10=(xa*pd).sum()
m01=(ya*pd).sum()
xm,ym=m10/m00, m01/m00
u20=(xa*xa*pd).sum()-xm*m10
u02=(ya*ya*pd).sum()-ym*m01
u11=(xa*ya*pd).sum()-ym*m10
tn2=2*u11/(u20-u02)
ang=arctan(tn2)/2.0
if u02>u20:
ang=ang-pi/2
xmean,ymean=X.mean(),Y.mean()
Xr = cos(ang) * (X-xmean) - sin(ang) * (Y-ymean) +xmean
Yr = sin(ang) * (X-xmean) + cos(ang) * (Y-ymean) +ymean
t=_polyMask(Xr,Yr,rs)
m00=t.sum()
m10=(xa*t).sum()
m01=(ya*t).sum()
xm,ym=m10/m00, m01/m00
u20=(xa*xa*t).sum()-xm*m10
u02=(ya*ya*t).sum()-ym*m01
u11=(xa*ya*t).sum()-ym*m10
aa=((16*u20**3)/(pi2*u02))**(1/8)
bb=((16*u02**3)/(pi2*u20))**(1/8)
##ellispe params
a,b=max([aa,bb]),min([aa,bb])
b=t.sum()/(pi*a)
ecc=sqrt(a**2-b**2)
theta=ang
ellArea=pi*a*b
print "Ploygon_area - ellispe_area = ",round(ellArea-t.sum(),3)
#print time.time()-t1
if showFig:
p.grid(True)
p.show()
#xee,yee=b/rs*cos(0),a/rs*sin(0)
#b3,a3=xee*max([Xmax,Ymax])/cos(theta),yee*min([Xmax,Ymax])/sin(theta)
#print b3/a3, b/a
return a/rs,b/rs,ecc,theta
def elltest(scale=0.8,off=0.2):
#generate an example, random, non-self-intersecting polygon.
#This is done by first generating
#it in polar coordinates and than translating it
#to cartesian.
Theta1,R1=linspace(0,2*pi,30),rand(30)*scale+off
X1,Y1=R1*cos(Theta1),R1*sin(Theta1)
X1=append(X1,X1[0])
Y1=append(Y1,Y1[0])
p.plot(X1,Y1,".-",ms=10)
a2,b2,ecc2,alpha2=ellfit(X1,Y1,showFig=False)
Xe,Ye=ellipse(b2,a2,-alpha2,X1.mean(),Y1.mean(),Nb=40)
p.plot(Xe,Ye,"r.-")
p.grid(True)
p.show()
pass
if __name__ == '__main__':
elltest()
Examples




Note 1
The algorithm has the following limitations:- The polygon must not be self intersecting.
- Polygon must be closed (last point must match the first one).
- Polygon must be centered in (0,0)
- Polygon must be within rectangle <-1,1>x<-1,1>
Note 2
This algorithm could be modified to overcome the above (Note 1) limitations. However, at the very moment I don't have time to do this. Hope this is not a problem.Note 3
Searching the Internet for an ellipse fitting script for Python I found fitellipse.py. It is very useful, however, sometimes I got an Exception that its not possible to fit an ellipse. The algorithm presented here seems to work good. In case the ellipse cannot be fitted try to increase rs attribute of ellfit function. The default value is 50 (rs=50).Note 3
This script is in ellipsefit.py.Tuesday, December 02, 2008
Python: use Cython to make it faster
Just as an example, I present some function that I translated to Cython. I also show how big the change in speed of my application was. The function is a part of an image processing application and it analyzes a given image on pixel by pixel basis. There are four nested loops! in this function. For this reason the function is very time consuming. The original code was:
def calculateVar(self): N=self.N;
I=self.ima
[ans,xoffs,yoffs,dists]=self.getSearchRegion()
noOfAngles=int(ans.shape[0])
self.ROTs=zeros(xoffs.shape)
diffMtx_size=N*N/self._jump/self._jump
diffMtx=zeros(diffMtx_size,dtype=int)
for ai in range(noOfAngles):
for offi in range(xoffs.shape[1]+0):
diffMtx.fill(0)
ind=0;
xoff=xoffs[ai,offi]
yoff=-yoffs[ai,offi]
for y1 in range(0,I.shape[0],self._jump):
for x1 in range(0,I.shape[1],self._jump):
x2=x1+xoff
y2=y1+yoff
if x2>=N or y2>=N or x2<0 ind="ind+1" diffmtx2="diffMtx[diffMtx">0]
self.ROTs[ai,offi]=diffMtx2.var()
This function was change to the following one:
def calculateVar(self):
N=self.N;
I=self.ima
I2=array(I,dtype=int)
[ans,xoffs,yoffs,dists]=self.getSearchRegion()
#this is part in Cython!
self.ROTs=loopcore.loopcore(I2,ans.shape[0],xoffs.shape[1],
array(xoffs,dtype=int),array(yoffs,dtype=int),
N, self._jump)where loopcore is a Cython module loopcore.pyx as follows:
import numpy as np
cimport numpy as np
DTYPE = np.int
ctypedef np.int_t DTYPE_t
ctypedef np.float_t DTYPE_t2
cdef inline int int_abs(int a, int b): return abs(a-b)
def loopcore(np.ndarray[DTYPE_t, ndim=2] I,int noOfAngles,
int noOfpixels, np.ndarray[DTYPE_t, ndim=2] xoffs,
np.ndarray[DTYPE_t, ndim=2] yoffs,
int N, int jump ):
cdef int y1,x1,x2, y2, ind,ai,offi, xoff,yoff,array_size
array_size=N*N/jump/jump
cdef np.ndarray[DTYPE_t, ndim=1] p= np.zeros(array_size, dtype=DTYPE)
cdef np.ndarray[DTYPE_t, ndim=1] p2= np.zeros(0, dtype=DTYPE)
cdef np.ndarray[DTYPE_t2, ndim=2] ROTs= np.zeros([noOfAngles,noOfpixels], dtype=np.float)
for ai in range(noOfAngles):
for offi in range(noOfpixels):
p.fill(0)
ind=0
xoff=xoffs[ai,offi]
yoff=-yoffs[ai,offi]
for y1 in range(0,N,jump):
for x1 in range(0,N,jump):
x2=x1+xoff
y2=y1+yoff
if x2>=N or y2>=N or x2<0 ind="ind+1" p2="p[p">0]
ROTs[ai,offi]=p2.var()
return ROTs
The gain in speed was huge. Execution of this function in Python takes about 2.10 min, while using Cython it takes about 0.05 min, i.e. code in Cython is 40+ times faster than that in Python. I'm sure that it can be made even faster than that!
Note 1
I noticed that the more variables is defined (cdef, int, float, ...) the greater the gain in speed is achieved.
Note 2
In Ubuntu 8.04 and 8.10 I compiled the Cython pyx files without any problems using the following command:
cython loopcore.pyx gcc -shared -pthread -fPIC -fwrapv -O2 -Wall -fno-strict-aliasing -I/usr/include/python2.5 -o loopcore.so loopcore.cNote 3
When running any python code from this post, remember to correct code indentations.
Wednesday, November 26, 2008
Python: draw an ellipse
The following Python script draws an ellipse.

from numpy import linspace
from scipy import pi,sin,cos
def ellipse(ra,rb,ang,x0,y0,Nb=50):
'''ra - major axis length
rb - minor axis length
ang - angle
x0,y0 - position of centre of ellipse
Nb - No. of points that make an ellipse
based on matlab code ellipse.m written by D.G. Long,
Brigham Young University, based on the
CIRCLES.m original
written by Peter Blattner, Institute of Microtechnology,
University of
Neuchatel, Switzerland, blattner@imt.unine.ch
'''
xpos,ypos=x0,y0
radm,radn=ra,rb
an=ang
co,si=cos(an),sin(an)
the=linspace(0,2*pi,Nb)
X=radm*cos(the)*co-si*radn*sin(the)+xpos
Y=radm*cos(the)*si+co*radn*sin(the)+ypos
return X,Y
def test():
import pylab as p
fig = p.figure(figsize=(5,5))
p.axis([-3,3,-3,3])
#eg 1
X,Y=ellipse(2,1,pi*2.0/3.0,0,1)
p.plot(X,Y,"b.-",ms=1) # blue ellipse
#eg 2
X,Y=ellipse(2,0.2,pi/3.0,1,1)
p.plot(X,Y,"r.-",ms=1) # red ellipse
#eg 3
X,Y=ellipse(1,1,pi/3.0,-1,1,Nb=16)
p.plot(X,Y,"g.-",ms=1) # green ellipse
p.grid(True)
p.show()
if __name__ == '__main__':
test()
Note
The script is in ellipse.py.Wednesday, November 05, 2008
Python: os.system returns 32512
The os.system() function executes operating systems's command. When the functin returns code 32512, it means that the command has not been found. One way to make it work, is to use the full path to the command.
En example:
En example:

Labels:
Python
Subscribe to:
Posts (Atom)

